NucleoMag kit for clean up and size selection of NGS library prep reactions
*taxes and shipping not included
Clean up and size selection of Next Generation Sequencing library preparation reactions
| Application | Clean up and size selection for NGS library preps |
| Selling unit | 5 mL, 50 mL, 500 mL |
| Target | Clean up |
| CE certified | No, research use only |
| Technology | Magnetic bead technology |
| Brand | NucleoMag |
| Format | Magnetic beads |
| Handling | Magnetic separation |
| Automated use | Yes |
| Sample material | Reaction mixtures from NGS library kits |
| Sample amount | 7.5 pg−5 µg nucleic acids in NGS reaction mixtures |
| Fragment size | 150 bp−approx. 800 bp |
| Typical recovery | ≥ 80 % |
| Elution volume | 10−100 µL |
| Preparation time | 40−120 min/96 preps |
| Typical downstream application | Next Generation Sequencing |
| Storage temperature | 2–8 °C / 36–46 °F |
| Shelf life (from production) | 24 Month(s) |
| Hazardous material | No |

Magnetic bead-based DNA and RNA Clean-up
NucleoMag NGS Clean-up and Size Select
Most sequencing platforms for library preparations require enzymatic reaction Clean-up and fragment size selection. NucleoMag NGS Clean-up and Size Select enables Clean-up reactions, as well as single or double sided size selection, to recover the fragment lengths which are needed in your specific application. Simply dilute the magnetic beads to utilize the tunable size selection feature. Dilution ratios are similar to most other comparable magnetic bead products in the market, allowing for the NucleoMag NGS Clean-up and Size Select kit to be used with your current protocols.

Product at a glance
| Technology | Magnetic beads |
| Applications | Size selection for next-generation-sequencing workflows PCR Clean-up RNA or cDNA Clean-up after purifications or enzymatic reactions (i. e. IVT or DNA digestion) |
| Recovery | 70 – 100 % |
| Elution volume | 10 – 100 μL |
| Shelf-life (from production) | 24 months |
| Product sizes | 5 mL, 50 mL, 500 mL |

Schematic Workflow Overview
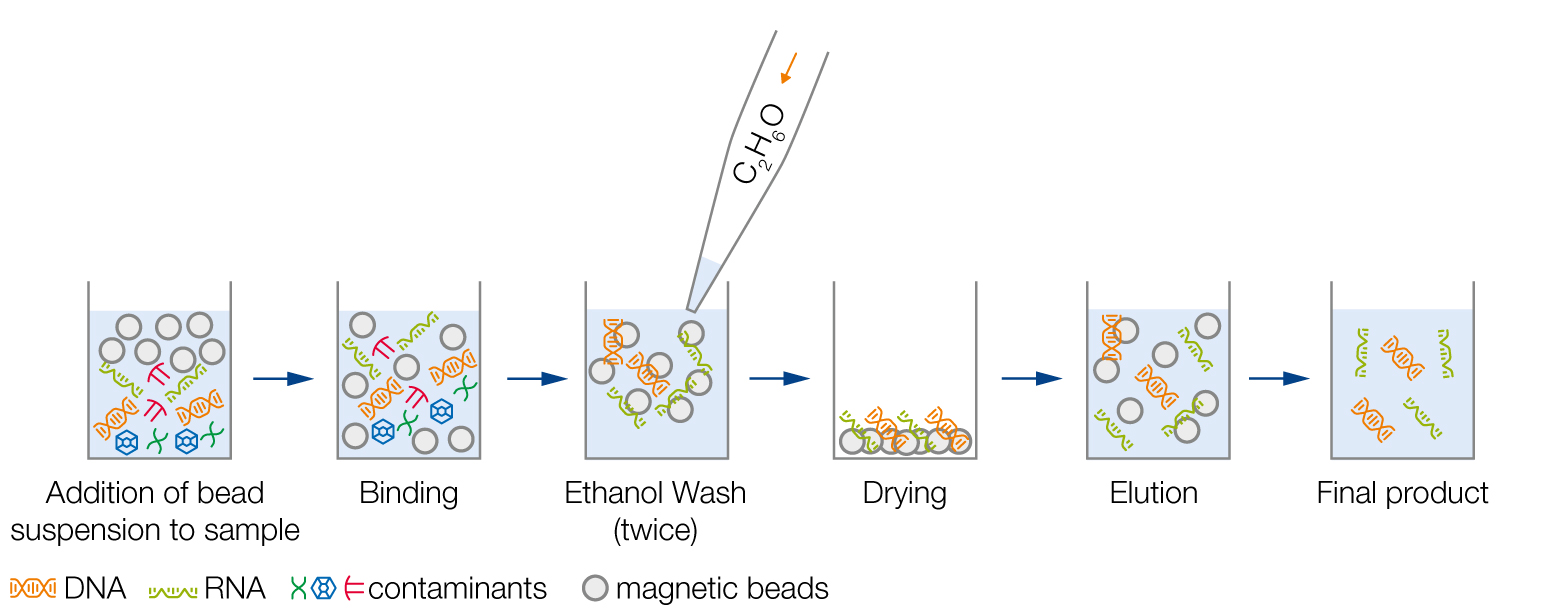
1. Add bead suspension to your DNA or RNA solution
2. Incubation of magnetic beads with sample for proper binding
3. Removal of contaminants by ethanol washes
4. Dry magnetic beads to allow evaporation of ethanol traces
5. Elution of DNA or RNA
6. Transfer of the final product to a new plate/tube
One Clean-up kit
for all samples!
fghfghgf
Use your
current protocol.
No changes
needed.
1. Add bead suspension to your DNA or RNA solution
2. Incubation of magnetic beads with sample for proper binding
3. Removal of contaminants by ethanol washes
4. Dry magnetic beads to allow evaporation of ethanol traces
5. Elution of DNA or RNA
6. Transfer of the final product to a new plate/tube
One Clean-up kit
for all samples!
fghfghgf
Use your
current protocol.
No changes
needed.
Application data – DNA Clean-Up and Size Selection
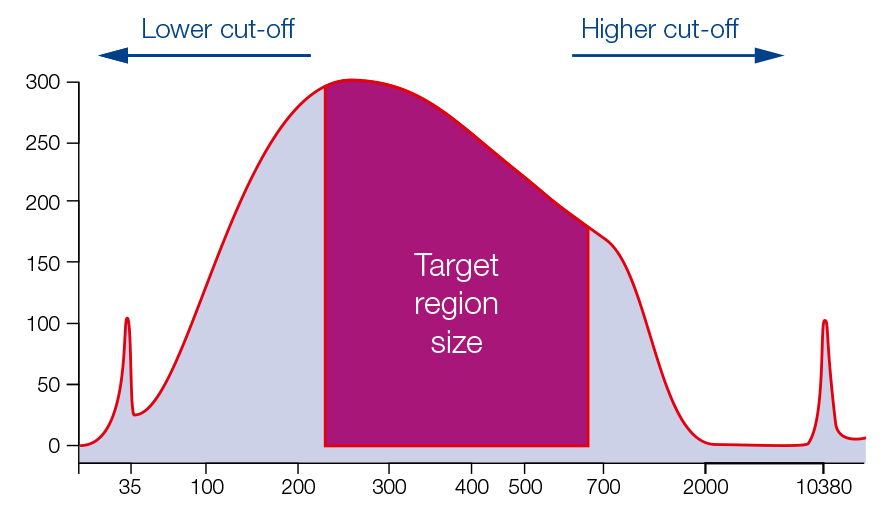
Size selection of fragment mix
For single sided size selection (left or right), the sample is mixed with the beads in predetermined ratios for the desired exclusion of smaller or larger sized fragments. For the double sided size selection, two binding steps are performed to exclude larger fragments above the cut-off and smaller fragments below the lower cut-off.
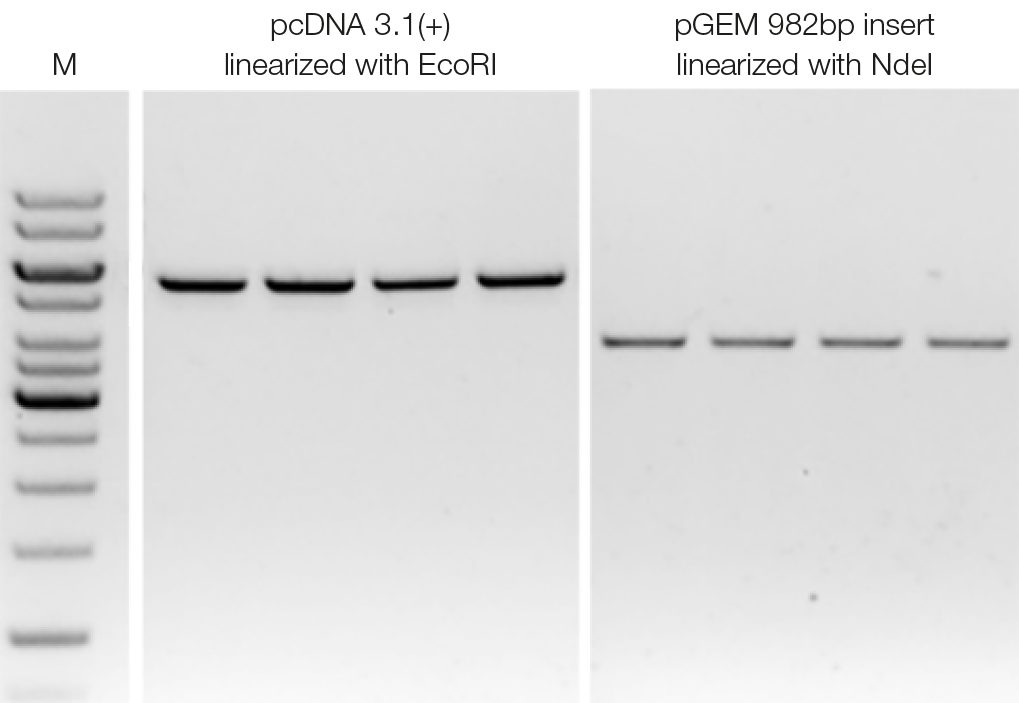
Reliable reproducibility in DNA clean-up procedures
To assess reproducibility of DNA clean-up, EcoRI-linearized pcDNA 3.1(+) and Ndel-linearized pGEM inserts were purified with four replicates each. For DNA purification, a sample to NGS clean-up suspension-ratio of 1.0 was used. The parallel Clean-up experiments show high reproducibility and recovery of the target DNA.
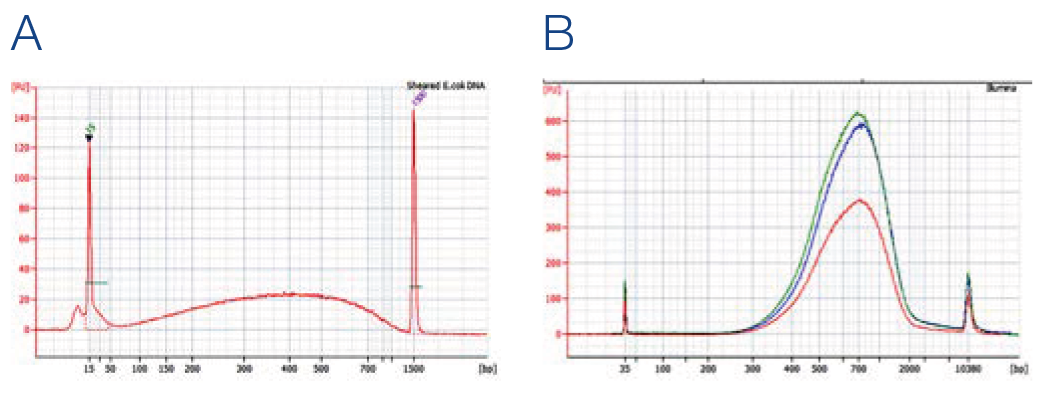
Fragment size analysis of prepared NGS libraries
NGS libraries were prepared using Truseq* DNA PCR Free kit from Illumina (red), AMPure* XP (blue) and NucleoMag NGS Clean-up and Size Select (green) for Clean-up and size selection steps. (A) Input DNA, 1 μg sheared E. coli DNA. (B) Size distribution of DNA Fragments after library preparation as input for sequencing with expected fragment size of 650 bp (insert + adapters).
Data kindly provided by TAKARA BIO INC.
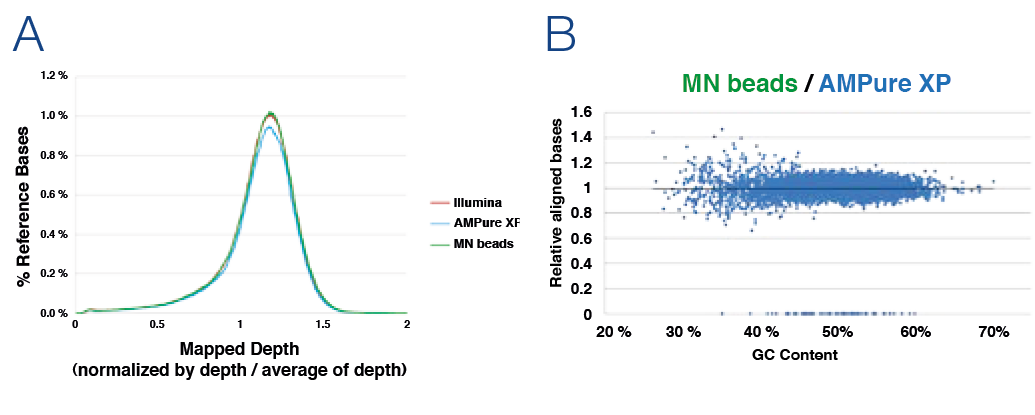
Sequencing analysis
In order to compare the sequencing performance after library preparation generation different clean-up and size selection products were tested. Performed tests included the distribution of genome sequence coverage (depth of genome base coverage) and effect on GC content on sequence coverage (A) No significant difference in mapped depth of genome base coverage. The constant depth indicates low bias. (B) Horizontal plot pattern of MN beads compared to AMPure* XP beads. After the summary of aligned read bases within a 0.5 kb genome region, the two samples have been normalized / matched resulting in a horizontally pattern showing the similarity of the two samples.
Data kindly provided by TAKARA BIO INC.
Cited in more than 50 publications
The NucleoMag NGS Clean-up and Size Select has been cited in more than 50 peer-reviewed publications. The kit has been used for PCR Clean-ups as well as in next generation sequencing workflows.
Application data – RNA Clean-Up
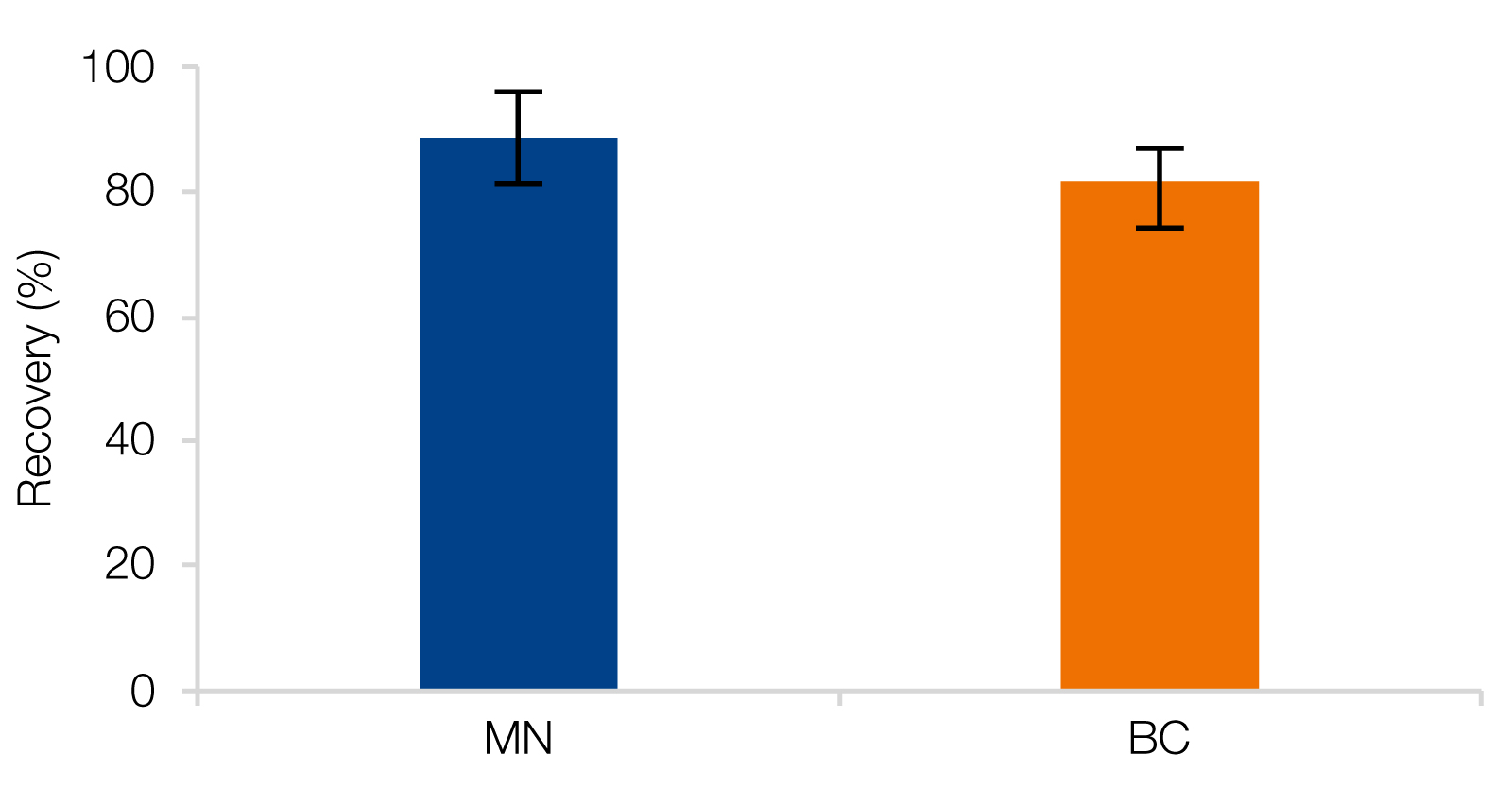
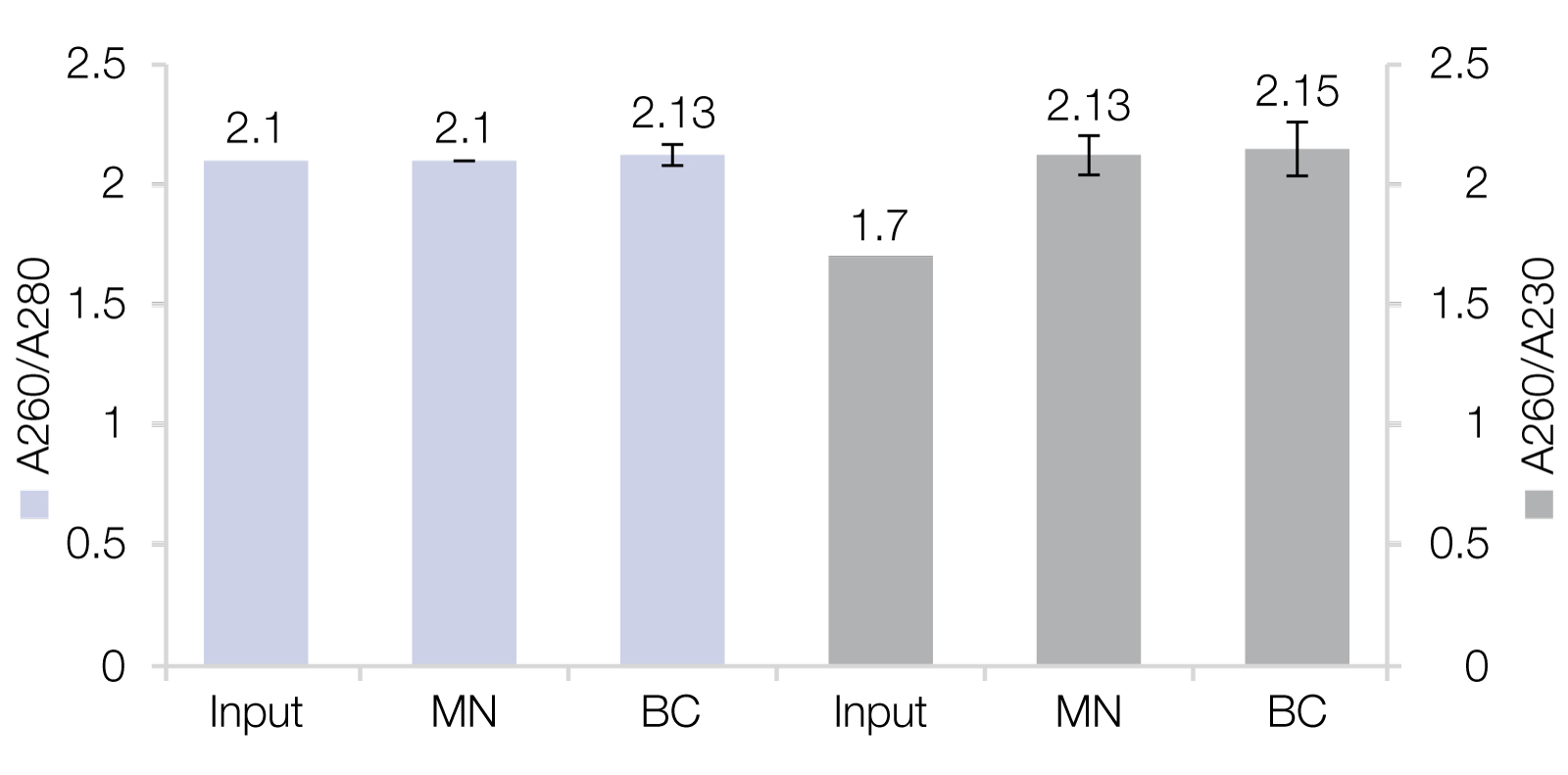
RNA Clean-Up with reliable recovery rates as well as high purities
A) MACHEREY‑NAGEL’s NucleoMag NGS Clean-up and Size Select beads as well as RNAClean* XP beads from Beckman Coulter (BC) were used for Clean-up of RiboRuler High Range Ladder, resulting in recovery rates of 88.6 ± 7.4 % (MN) and 81.5 ± 5.4 % (BC). B) RNA isolated from HeLa cells were used as input for Clean-up reactions which generated improved purities.
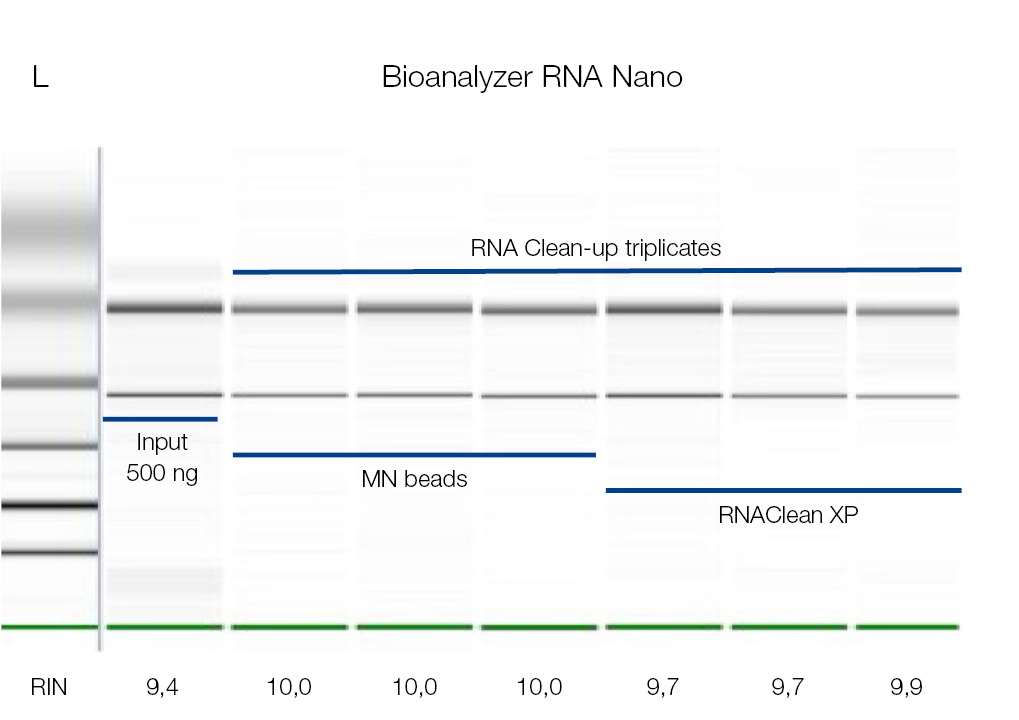
High-integrity RNA after Clean-up with NucleoMag NGS Clean-Up and Size Select kit
RNA samples were cleaned-up using the NucleoMag NGS Clean-up and Size Select kit (MACHEREY‑NAGEL) as well as RNAClean* XP beads (Beckman Coulter). 500 ng RNA was used as starting material. The RNA was eluted in 25 μL and analyzed on Agilent’s Bioanalyzer 2100. No significant difference in RNA integrity has been observed for used products, as integrity values after clean-up are comparable to the input sample (RIN ≥ 9,4).
NucleoMag NGS Clean-Up and Size Select kit is suitable for Clean-ups of RNA transcripts after in vitro transcription reactions
Riboprobe* System – T7 has been used for in vitro transcription of single-stranded RNA using a pGEM* vector. NucleoMag NGS Clean-up and Size Select and RNAClean* XP were used for Clean-up of IVT reactions.
| Product | ng/μL | A260 / A280 | A260 / A230 |
|---|---|---|---|
| MN Beads | 14.4 ± 2.1 | 2.3 ± 0.1 | 2.5 ± 0.7 |
| RNAClean* XP | 14.4 ± 1.6 | 2.3 ± 0.1 | 2.2 ± 0.8 |
Technical data & contact information
Please follow the link below or read the product broschure for more information on technical data, specifications and related products.
Contact the experts from MACHEREY‑NAGEL technical support!
Email: support@mn-net.com
Phone: +49 (0) 2421 969 333
| Product | ng/μL | A260 / A280 | A260 / A230 |
|---|---|---|---|
| MN Beads | 14.4 ± 2.1 | 2.3 ± 0.1 | 2.5 ± 0.7 |
| RNAClean* XP | 14.4 ± 1.6 | 2.3 ± 0.1 | 2.2 ± 0.8 |
* Trademarks: AMPure (Beckman Coulter, Inc.), RNAClean (Beckman Coulter, Inc.), Riboprobe (Promega Corp.), pGEM (Promega Corp.) and Truseq (Illumina)
 NucleoSpin Gel and PCR Clean-up, Mini kit for gel extraction or PCR clean up
NucleoSpin Gel and PCR Clean-up, Mini kit for gel extraction or PCR clean up